Open-source data platform for biology biology
Manage data & analyses with an open-source Python framework.
Collaborate across dry and wet lab in a distributed data hub.
Start on your laptop. Deploy anywhere.
Unified access
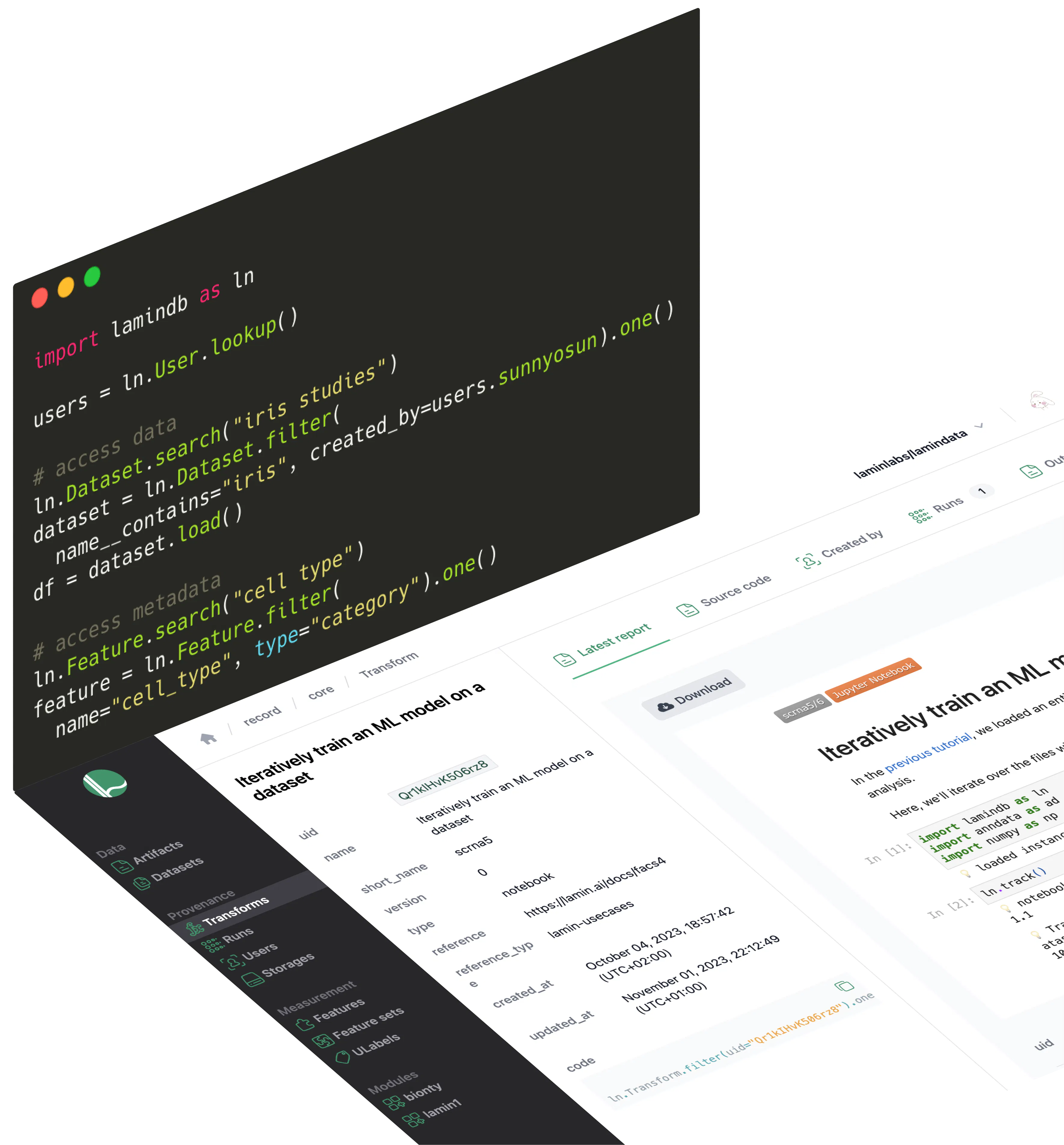
Unify access to data & metadata
No more wrangling queries across fragmented data lakes & ELN/LIMS systems.
Use one API & UI to access & manage data and metadata. Bridge data artifacts and warehousing. Plug in custom schemas and manage migrations with ease.
Data flow
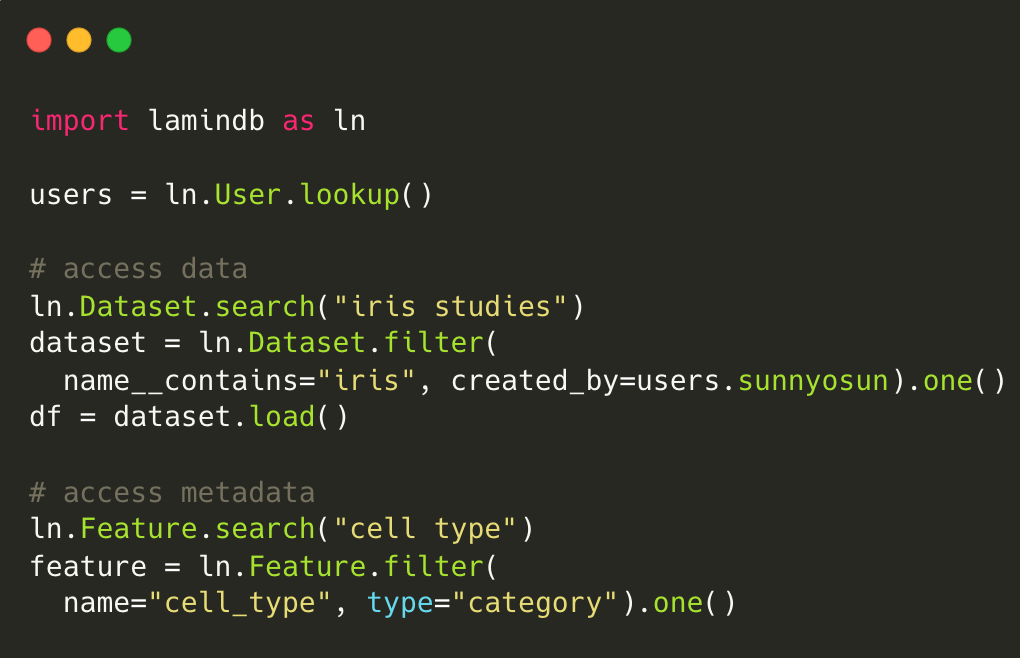
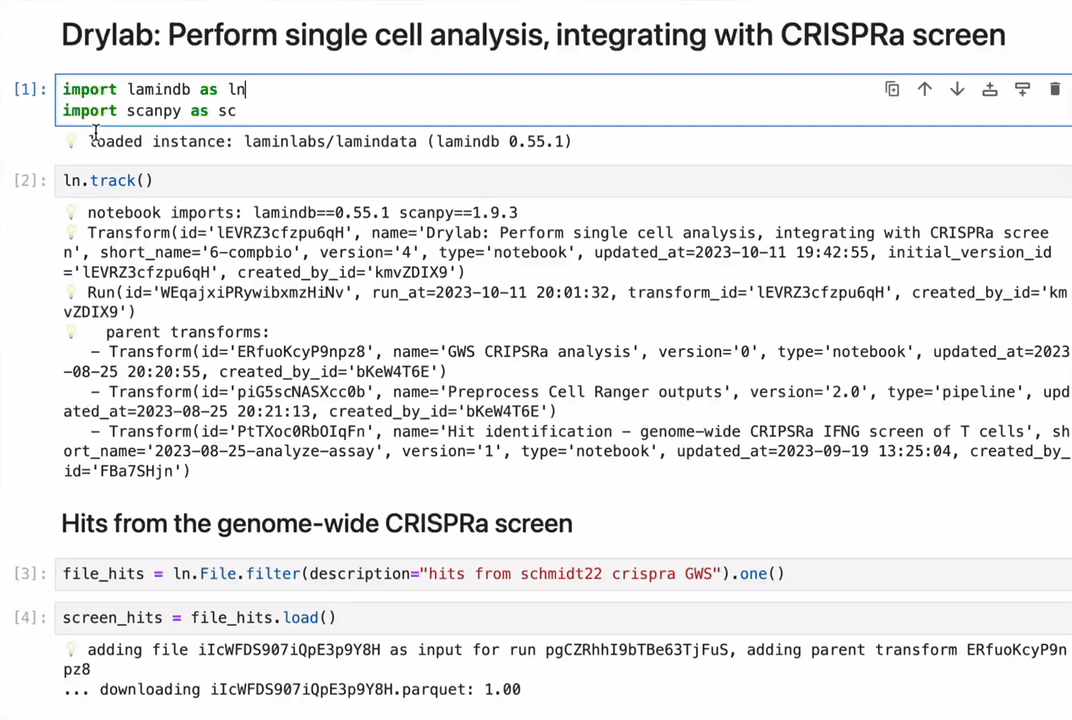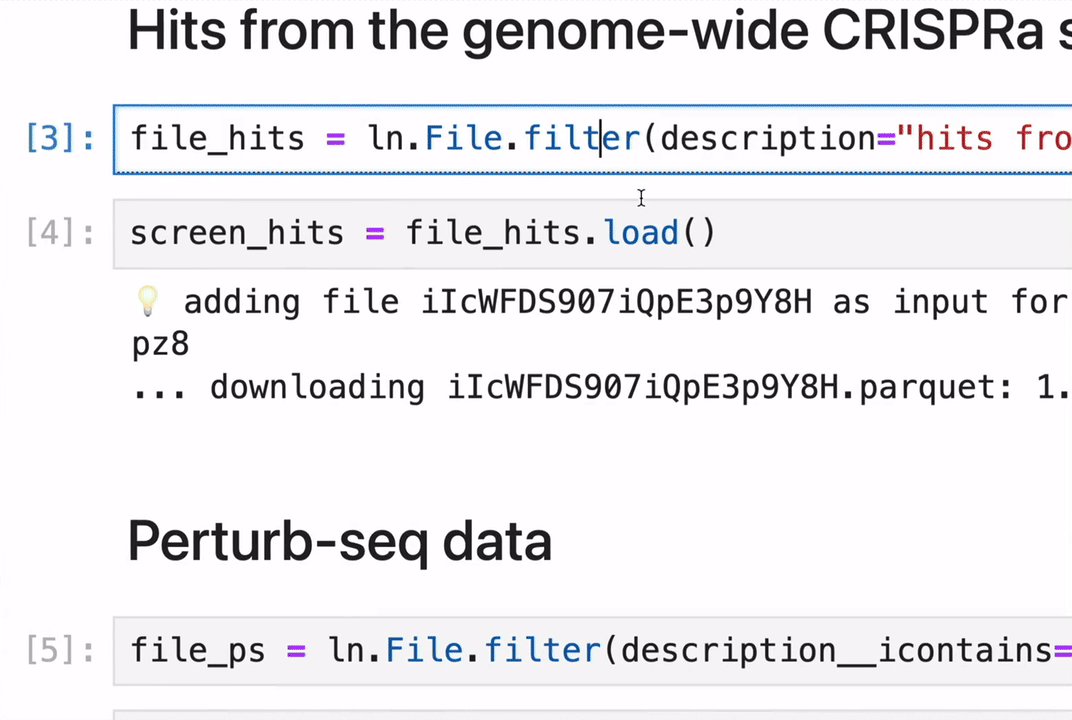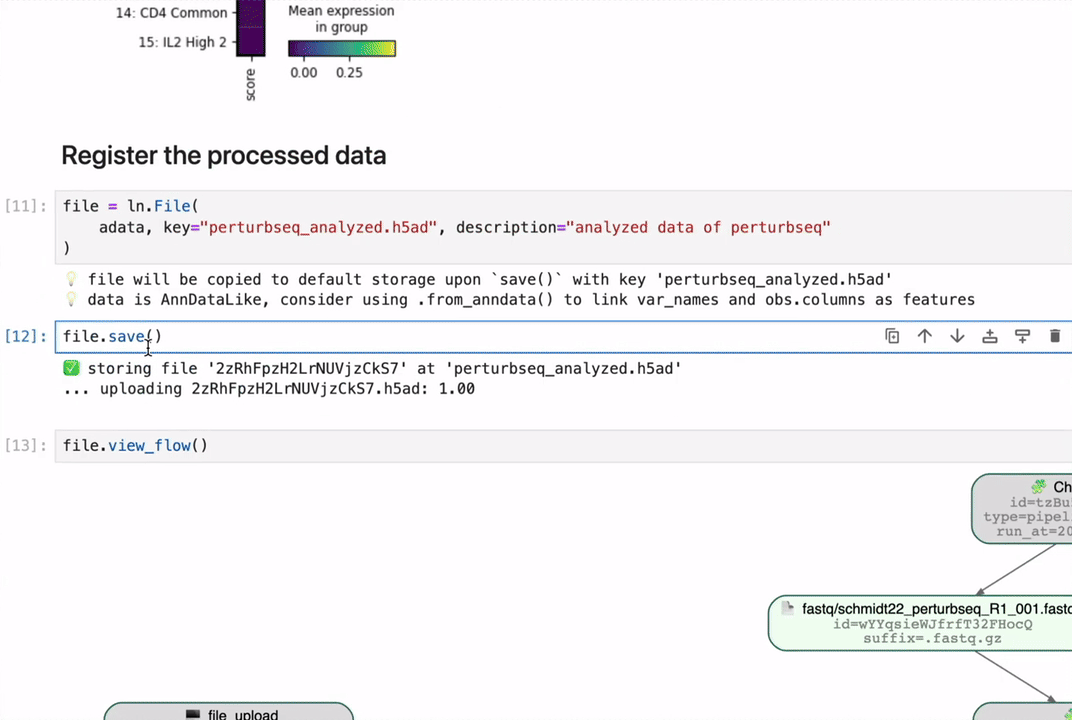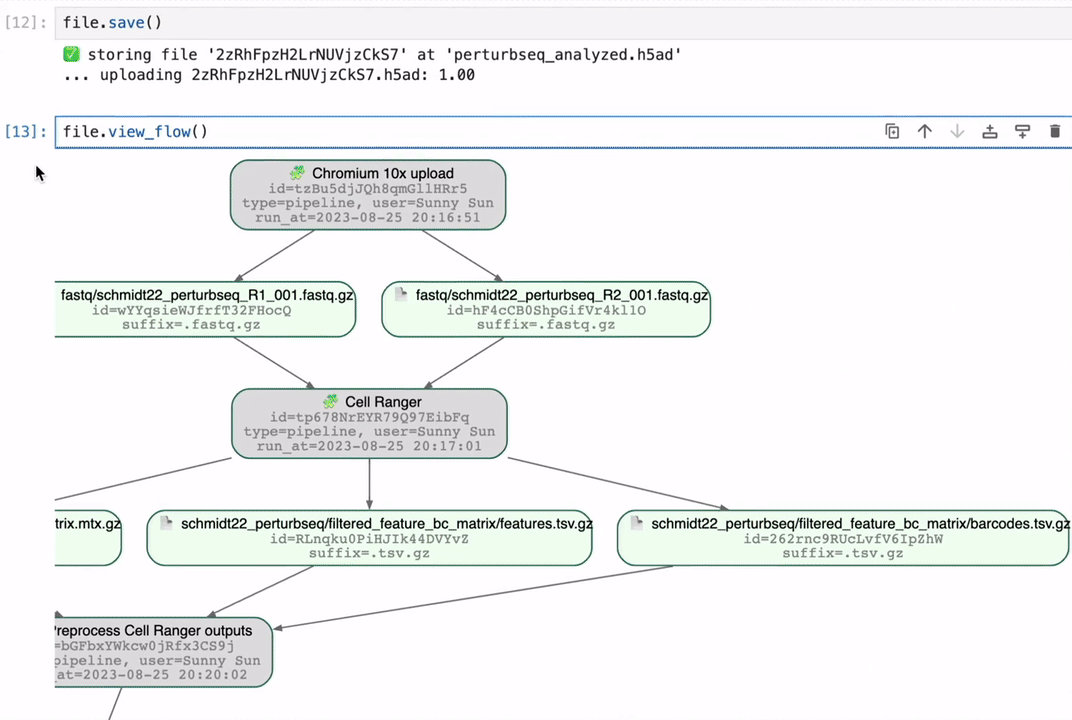
Track data lineage across notebooks, pipelines & UI
No more guessing where an analytical result came from.
Rely on and understand how data transforms across notebooks, pipelines & UI. Integrate with workflow managers like redun, Nextflow, and Snakemake.
Biological registries
Manage registries for experimental metadata & ontologies
No more confusion about fundamental entities.
Manage Gene, Protein, CellType, CellLine, Tissue, Disease, etc. records in ontology-aware registries. Import data from public ontologies with ease. Navigate entities with auto-completion.
Data validation
Validate, standardize & annotate
No more typos, duplicates and multiple names for the same thing.
Validate and standardize with one-line API calls. Annotate your data with untyped or typed labels & features.
Data share
Organize data across a mesh of instances
No more never-ending alignment on schemas across teams.
Instantly create & load LaminDB instances like git repositories and collaborate on them in LaminHub. Zero-copy transfer data across instances.
Process
Learn & warehouse in canonical iterations
No more monstrous notebooks & scripts that do everything.
Use an atomic iterative process to consistently transform files into more useful representations: validated, queryable datasets and analytical insights.